Hieff Trans™ Liposomal Transfection Reagent
Description
Hieff Trans™ Liposomal Transfection Reagent is a versatile liposome transfection reagent, suitable for DNA, RNA, and oligonucleotide transfection, with high transfection efficiency for most eukaryotic cells. Its unique formula allows it to be added directly to the medium, and the presence of serum does not affect transfection efficiency, which reduces the damage to cells caused by serum removal. There is no need to remove the nucleic acid-Hieff Trans™ complex or replace it with a fresh medium after transfection, and it can also be removed after 4-6 hours.
Hieff Trans™ is supplied in sterile liquid form. Usually, for 24-well plate transfection, about 1.5 μL each time, 1 mL of Hieff Trans™ can do about 660 transfections; for a 6-well plate, about 6 μL each time, 1 mL of Hieff Trans™ can do about 660 transfections. 160 transfections.
Feature
- Exceptional Efficiency: Superior transfection performance in the broad range of cell lines, including transient transfection and stable transfection
- Wide Adaptability: Excellent transfection efficiency in a variety of cell lines and high levels of recombinant protein expression
- Low Toxicity: The activities of the diverse cells almost unaffected by transfection reagents from YEASEN
- Simple Operation: Proven efficacy in the presence of serum—eliminates the need to change media following transfection
- Cost Effective: Competitive transfection effect with more affordable prices
Application
- Cell Transfection
Specification
| Form | Liquid |
| Serum Compatible | Yes |
| Cell Type | Established Cell Lines |
| Sample Type | Plasmid DNA, Synthetic siRNA |
| Transfection Technique | Lipid-Based Transfection |
Components
| Components No. | Name | 40802ES02 | 40802ES03 | 40802ES08 |
| 40802 | Hieff TransTM;Liposomal Transfection Reagent | 0.5 mL | 1 mL | 5×1 mL |
Shipping and Storage
The product is shipped with ice packs and can be stored at 2-8ºC for one year. Do not freeze!
Figures
- Demonstration of Transfection Effect
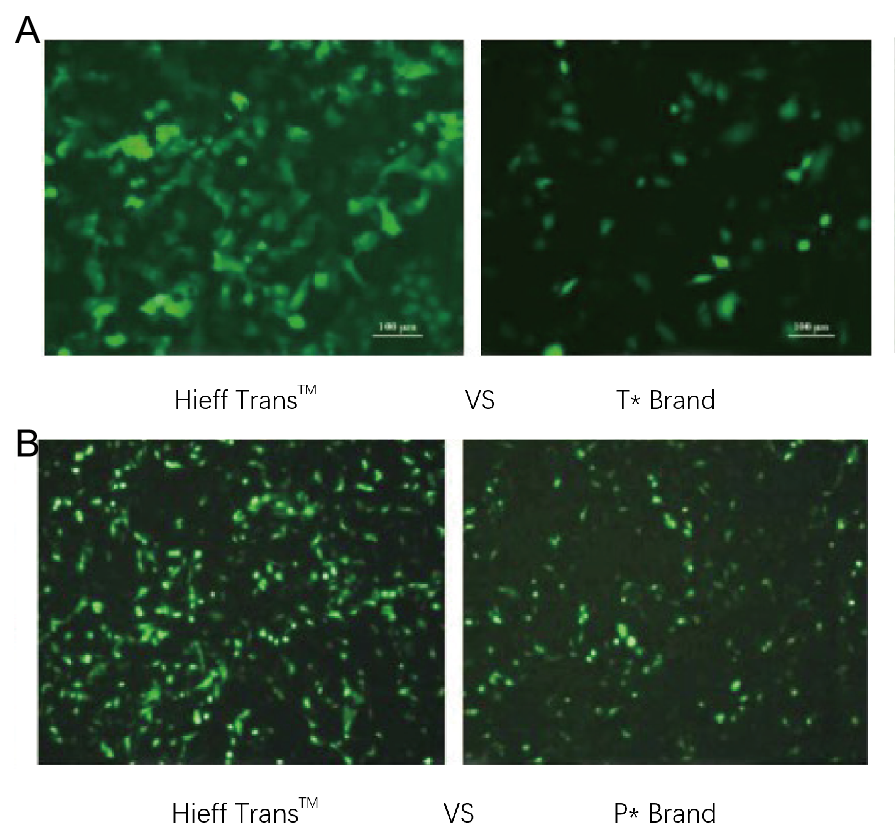
Figure 1. Hieff Trans™ Liposome Transfection Reagent outperforms the transfection reagent from the competitive brand(T* brand in Figure 1a, P* brand in Figure 1b).
Each reagent was used to transfect the target cell line(Hela cell line in Figure 1a, DF-1 cell line in Figure 1b)in a 96-well format. GFP expression was analyzed 48 hours posttransfection. Hieff Trans™ Liposome Transfection Reagent provided higher GFP transfection efficiency than the competitive brand products.
- Validated Cell Lines
| Product Name | Hieff TransTMLiposomal Transfection Reagent | |||||
| Validated Cell Lines | 293T | Calu 1 | HEK293 | HO1980 | N2A | TS |
| 293F | CHO-K1 | HEK293T | HUVEC | NCI-H1975 | U-87 | |
| 293FT | COS-7 | Hela | MCF10A | NIH-3T3 | Vero | |
| 3T3 | DF-1 | Hep2C | MCF-7 | Neuro-2a | WEHI | |
| A549 | H1299 | Hep3B | MDA-MB-231 | PC-12 | WRL-68 | |
| BV-2 | H520 | Hepa1-6 | MDA-MB-231-LM2-4175 | Raw264.7 | ||
| B50 | HaCaT | HepG2 | MDCK | SGC-7901 | ||
| C2C12 | HCT116 | HK2 | MEF | T47D | … | |
Figure 2. List of cell lines successfully transfected with Hieff Trans™ Liposomal Transfection Reagent (Under continuous update).

Cited from "Optogenetic control of RNA function and metabolism using engineered light-switchable RNA-binding proteins. Nat Biotechnol. 2022 Jan 3. doi: 10.1038/s41587-021-01112-1"

Cited from "Structural insight into BRCA1-BARD1 complex recruitment to damaged chromatin. Mol Cell. 2021 Jul 1;81(13):2765-2777.e6. doi: 10.1016/j.molcel.2021.05.010."
Cited from "UBQLN2-HSP70 axis reduces poly-Gly-Ala aggregates and alleviates behavioral defects in the C9ORF72 animal model. Neuron. 2021 Jun 16;109(12):1949-1962.e6. doi: 10.1016/j.neuron.2021.04.023."
Hieff Trans™ Liposomal Transfection Reagent FAQ
(1) Q: Can serum be present when preparing nucleic acid transfection reagent complex?
A: The presence of serum will affect the formation of liposomes. It is recommended to use serum-free medium (usually MEM medium) when preparing nucleic acid transfection reagent complexes.
(2) Q: Can the transfection reagent be frozen?
A: No. This reagent must be stored at 2-8 ℃, and care should be taken to avoid repeatedly opening the cap for a long time, as long-term opening of the cap will cause liposome oxidation and affect the transfection efficiency.
(3) Q: What should I pay attention to when using Hieff Trans™ Liposome Nucleic Acid Transfection Reagent?
A: 1) During the transfection operation, it is better that the cell confluence reaches 80%-95%, and the specific plating density is determined according to the situation of the cells;
2) Using high-purity DNA helps to obtain higher transfection efficiency;
3) DNA and transfection reagents are required to be diluted with serum-free medium when preparing transfection complexes;
4) Antibiotics cannot be added to the medium during transfection;
5) The DNA concentration and the amount of cationic liposome reagent should be optimized for the first use to obtain the maximum transfection efficiency. The ratio of DNA to transfection reagent is generally recommended to be 1:2-1:3.
(4) Q: Does it need to be terminated after transfection?
A: No need. Liposome complexes are stable for 6 hours. If the cell medium is not changed before transfection, in order to ensure the nutrients required for normal cell growth, it is necessary to change to a new medium after 4 to 6 hours. However, if the medium has been changed before transfection, it is not necessary to change the medium after liposome transfection.
(5) Q: What should I pay attention to if I want to improve the transfection efficiency?
A: a: The density of cells at the time of transfection is 90%-95%.
b: During transfection, use MEM serum-free medium for nucleic acid and liposome dilutions.
c: The medium can be changed 4-6h after transfection.
(6) Q: Can co-transfection of DNA and siRNA be performed? How's the effect?
A: Co-transfection can be performed, but it is recommended to perform separate transfection, and DNA transfection should be performed 6 hours after siRNA. If operated together, the siRNA transfection efficiency will be worse.
(7) Q: Can the transfection reagent be used for lentiviral packaging transfection?
A: Lentiviral packaging is possible, but the efficiency of lentiviral packaging is not necessarily related to the efficiency of transfection, but also related to the selection of packaging plasmids and the ratio between plasmids.
(8) Q: Can Hieff Trans™ Liposome Nucleic Acid Transfection Reagent be used for transfection of suspension cells?
A: Hieff Trans™ Liposome Nucleic Acid Transfection Reagent can be used for suspension cell transfection, see Protocol for details. In addition, we have also launched a transfection reagent specifically for suspension cells (Cat No. 40805, Liposome nucleic acid transfection reagent for suspension cells).
[1] Liu R, Yang J, Yao J, Zhao Z, He W, Su N, Zhang Z, Zhang C, Zhang Z, Cai H, Zhu L, Zhao Y, Quan S, Chen X, Yang Y. Optogenetic control of RNA function and metabolism using engineered light-switchable RNA-binding proteins. Nat Biotechnol. 2022 Jan 3. doi: 10.1038/s41587-021-01112-1. Epub ahead of print. PMID: 34980910. (IF:54.908)
[2] Zhou J, Chen P, Wang H, Liu H, Li Y, Zhang Y, Wu Y, Paek C, Sun Z, Lei J, Yin L. Cas12a variants designed for lower genome-wide off-target effect through stringent PAM recognition. Mol Ther. 2022 Jan 5;30(1):244-255. doi: 10.1016/j.ymthe.2021.10.010. Epub 2021 Oct 20. PMID: 34687846; PMCID: PMC8753454. (IF:11.454)
[3] Chen S, Cao X, Zhang J, Wu W, Zhang B, Zhao F. circVAMP3 Drives CAPRIN1 Phase Separation and Inhibits Hepatocellular Carcinoma by Suppressing c-Myc Translation. Adv Sci (Weinh). 2022 Mar;9(8):e2103817. doi: 10.1002/advs.202103817. Epub 2022 Jan 24. PMID: 35072355; PMCID: PMC8922094. (IF:16.808)
[4] Zhang Y, Yu X, Sun R, Min J, Tang X, Lin Z, Xie S, Li X, Lu S, Tian Z, Gu C, Teng L, Yang Y. Splicing factor arginine/serine-rich 8 promotes multiple myeloma malignancy and bone lesion through alternative splicing of CACYBP and exosome-based cellular communication. Clin Transl Med. 2022 Feb;12(2):e684. doi: 10.1002/ctm2.684. PMID: 35184390. (IF:11.492)
[5] Tang X, Deng Z, Ding P, Qiang W, Lu Y, Gao S, Hu Y, Yang Y, Du J, Gu C. A novel protein encoded by circHNRNPU promotes multiple myeloma progression by regulating the bone marrow microenvironment and alternative splicing. J Exp Clin Cancer Res. 2022 Mar 8;41(1):85. doi: 10.1186/s13046-022-02276-7. PMID: 35260179. (IF:11.161)
[6] Hua Z, Wei R, Guo M, Lin Z, Yu X, Li X, Gu C, Yang Y. YTHDF2 promotes multiple myeloma cell proliferation via STAT5A/MAP2K2/p-ERK axis. Oncogene. 2022 Mar;41(10):1482-1491. doi: 10.1038/s41388-022-02191-3. Epub 2022 Jan 24. PMID: 35075244. (IF:9.867)
[7] Liang Y, Lu Q, Li W, Zhang D, Zhang F, Zou Q, Chen L, Tong Y, Liu M, Wang S, Li W, Ren X, Xu P, Yang Z, Dong S, Zhang B, Huang Y, Li D, Wang H, Yu W. Reactivation of tumour suppressor in breast cancer by enhancer switching through NamiRNA network. Nucleic Acids Res. 2021 Sep 7;49(15):8556-8572. doi: 10.1093/nar/gkab626. PMID: 34329471; PMCID: PMC8421228. (IF:16.9)
[8] Dai L, Dai Y, Han J, Huang Y, Wang L, Huang J, Zhou Z. Structural insight into BRCA1-BARD1 complex recruitment to damaged chromatin. Mol Cell. 2021 Jul 1;81(13):2765-2777.e6. doi: 10.1016/j.molcel.2021.05.010. Epub 2021 Jun 7. PMID: 34102105. (IF:17.97)
[9] Zhang K, Wang A, Zhong K, Qi S, Wei C, Shu X, Tu WY, Xu W, Xia C, Xiao Y, Chen A, Bai L, Zhang J, Luo B, Wang W, Shen C. UBQLN2-HSP70 axis reduces poly-Gly-Ala aggregates and alleviates behavioral defects in the C9ORF72 animal model. Neuron. 2021 Jun 16;109(12):1949-1962.e6. doi: 10.1016/j.neuron.2021.04.023. Epub 2021 May 14. PMID: 33991504. (IF:17.17)
[10] Liang Y, Lu Q, Li W, Zhang D, Zhang F, Zou Q, Chen L, Tong Y, Liu M, Wang S, Li W, Ren X, Xu P, Yang Z, Dong S, Zhang B, Huang Y, Li D, Wang H, Yu W. Reactivation of tumour suppressor in breast cancer by enhancer switching through NamiRNA network. Nucleic Acids Res. 2021 Sep 7;49(15):8556-8572. doi: 10.1093/nar/gkab626. PMID: 34329471; PMCID: PMC8421228. (IF:16.9)
[11] Li T, Chen X, Qian Y, Shao J, Li X, Liu S, Zhu L, Zhao Y, Ye H, Yang Y. A synthetic BRET-based optogenetic device for pulsatile transgene expression enabling glucose homeostasis in mice. Nat Commun. 2021 Jan 27;12(1):615. doi: 10.1038/s41467-021-20913-1. PMID: 33504786; PMCID: PMC7840992. (IF:14.92)
[12] Pan Y, He X, Li C, Li Y, Li W, Zhang H, Wang Y, Zhou G, Yang J, Li J, Qu J, Wang H, Gao Z, Shen Y, Li T, Hu H, Ma H. Neuronal activity recruits the CRTC1/CREB axis to drive transcription-dependent autophagy for maintaining late-phase LTD. Cell Rep. 2021 Jul 20;36(3):109398. doi: 10.1016/j.celrep.2021.109398. PMID: 34289350. (IF:9.42)
[13] Liu H, Xing R, Ou Z, Zhao J, Hong G, Zhao TJ, Han Y, Chen Y. G-protein-coupled receptor GPR17 inhibits glioma development by increasing polycomb repressive complex 1-mediated ROS production. Cell Death Dis. 2021 Jun 12;12(6):610. doi: 10.1038/s41419-021-03897-0. PMID: 34120140; PMCID: PMC8197764. (IF:8.463)
[14] Fan Y, Wang J, Jin W, Sun Y, Xu Y, Wang Y, Liang X, Su D. CircNR3C2 promotes HRD1-mediated tumor-suppressive effect via sponging miR-513a-3p in triple-negative breast cancer. Mol Cancer. 2021 Feb 2;20(1):25. doi: 10.1186/s12943-021-01321-x. PMID: 33530981; PMCID: PMC7851937. (IF:27.403)
[15] Gu C, Wang Y, Zhang L, Qiao L, Sun S, Shao M, Tang X, Ding P, Tang C, Cao Y, Zhou Y, Guo M, Wei R, Li N, Xiao Y, Duan J, Yang Y. AHSA1 is a promising therapeutic target for cellular proliferation and proteasome inhibitor resistance in multiple myeloma. J Exp Clin Cancer Res. 2022 Jan 6;41(1):11. doi: 10.1186/s13046-021-02220-1. PMID: 34991674; PMCID: PMC8734095. (IF:11.161)
[16] Luo Q, Wu X, Zhao P, Nan Y, Chang W, Zhu X, Su D, Liu Z. OTUD1 Activates Caspase-Independent and Caspase-Dependent Apoptosis by Promoting AIF Nuclear Translocation and MCL1 Degradation. Adv Sci (Weinh). 2021 Feb 8;8(8):2002874. doi: 10.1002/advs.202002874. PMID: 33898171; PMCID: PMC8061361. (IF:15.84)
[17] Luo Q, Wu X, Nan Y, Chang W, Zhao P, Zhang Y, Su D, Liu Z. TRIM32/USP11 Balances ARID1A Stability and the Oncogenic/Tumor-Suppressive Status of Squamous Cell Carcinoma. Cell Rep. 2020 Jan 7;30(1):98-111.e5. doi: 10.1016/j.celrep.2019.12.017. PMID: 31914402. (IF:9.42)
[18] Sun X, Peng X, Cao Y, Zhou Y, Sun Y. ADNP promotes neural differentiation by modulating Wnt/β-catenin signaling. Nat Commun. 2020 Jun 12;11(1):2984. doi: 10.1038/s41467-020-16799-0. PMID: 32533114; PMCID: PMC7293280. (IF:14.911)
[19] Yang X, Wang H, Xie E, Tang B, Mu Q, Song Z, Chen J, Wang F, Min J. Rewiring ERBB3 and ERK signaling confers resistance to FGFR1 inhibition in gastrointestinal cancer harbored an ERBB3-E928G mutation. Protein Cell. 2020 Dec;11(12):915-920. doi: 10.1007/s13238-020-00749-z. PMID: 32632529; PMCID: PMC7719122. (IF:14.872)
[20] Chen, T., Chen, Y., Chen, H. et al. Dual-enzyme-propelled unbounded DNA walking nanomachine for intracellular imaging of lowly expressed microRNA. Nano Res. 12, 1055–1060 (2019). https://doi.org/10.1007/s12274-019-2344-5 (IF:8.21)
[21] Zhang X, Qi Z, Yin H, Yang G. Interaction between p53 and Ras signaling controls cisplatin resistance via HDAC4- and HIF-1α-mediated regulation of apoptosis and autophagy. Theranostics. 2019 Jan 30;9(4):1096-1114. doi: 10.7150/thno.29673. PMID: 30867818; PMCID: PMC6401400. (IF:8.12)
[22] Zou Y, Wang A, Shi M, Chen X, Liu R, Li T, Zhang C, Zhang Z, Zhu L, Ju Z, Loscalzo J, Yang Y, Zhao Y. Analysis of redox landscapes and dynamics in living cells and in vivo using genetically encoded fluorescent sensors. Nat Protoc. 2018 Oct;13(10):2362-2386. doi: 10.1038/s41596-018-0042-5. PMID: 30258175; PMCID: PMC6714056. (IF:13.49)
[23] Zhang K, Zhao X, Chen X, Wei Y, Du W, Wang Y, Liu L, Zhao W, Han Z, Kong D, Zhao Q, Guo Z, Han Z, Liu N, Ma F, Li Z. Enhanced Therapeutic Effects of Mesenchymal Stem Cell-Derived Exosomes with an Injectable Hydrogel for Hindlimb Ischemia Treatment. ACS Appl Mater Interfaces. 2018 Sep 12;10(36):30081-30091. doi: 10.1021/acsami.8b08449. Epub 2018 Aug 29. PMID: 30118197. (IF:8.09)
[24] Hao H, Hu S, Chen H, Bu D, Zhu L, Xu C, Chu F, Huo X, Tang Y, Sun X, Ding BS, Liu DP, Hu S, Wang M. Loss of Endothelial CXCR7 Impairs Vascular Homeostasis and Cardiac Remodeling After Myocardial Infarction: Implications for Cardiovascular Drug Discovery. Circulation. 2017 Mar 28;135(13):1253-1264. doi: 10.1161/CIRCULATIONAHA.116.023027. Epub 2017 Feb 2. PMID: 28154007. (IF:18.881)
Catalog No.:*
Name*
phone Number:*
Lot:*
Email*
Country:*
Company/Institute:*

