Hifair® Precision sgRNA Synthesis Kit
Product description
CRISPR/Cas9 is a technique in which RNA-directed Cas nuclease to make specific DNA modifications to targeted genes, which is an acquired immune defense mechanism evolved by bacteria and archaebacteria in response to attacks by phages and foreign plasmids. In this system, the Cas9 nuclease is guided with artificially optimized single guide RNA (sgRNA) to cut double-stranded DNA (dsDNA) at the target site causing DNA breaks. DNA is repaired with the non-homologous end joining or Homology directed repair mechanism in vivo, resulting in frameshift mutation, replacement or deletion of genes.
HifairTM Precision sgRNA Synthesis Kit uses a T7 RNA polymerase mixture to efficiently transcribe spCas9 sgRNA. The kit contains a specific sequence that is recognized by the Cas9 protein and acts as a scaffold. A portion of the sequence can be partially overlapped with the target specific sequence designed by the user. The complementary strands are filled by DNA polymerase to generate dsDNA templates for transcription. Using its supplied reagent and user-designed specific DNA sequences, the kit can obtain 20-100 μg of functional sgRNA within 4 hours in a single tube reaction.
Components
|
Components No. |
Name |
11355ES05 |
11355ES25 |
11355ES50 |
11355ES60 |
|
11355-A |
10×sgRNA Enzyme Mix |
10 μL |
50 μL |
100 μL |
200 μL |
|
11355-B |
10×sgRNA Reaction Buffer |
10 μL |
50 μL |
100 μL |
200 μL |
|
11355-C |
2×Canace Enzyme Mix |
62.5 μL |
312.5 μL |
625 μL |
1.25 mL |
|
11355-D |
Scaffold Template |
6.25 μL |
31.25 μL |
62.5 μL |
125 μL |
|
11355-E |
NTPs (25 mM each) |
40 μL |
200 μL |
400 μL |
800 μL |
|
11355-F |
Control sgRNA Oligo (10 μM) |
2 μL |
10 μL |
20 μL |
40 μL |
Storage
This product should be stored at -25~-15℃ for 2 years.
Instructions
- Synthesis principle
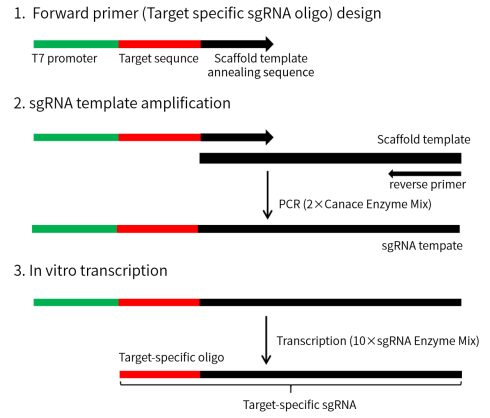
2. Operation procedure
2.1 Forward primer (Target specific sgRNA oligo) design
- The 5' end of the primer: T7 promoter sequence with protection base (20nt:TTCTAATACGACTCACTATA).
- Transcription start site: Add 0~2 G, the number of G added is determined by the 5 'end of the target sequence, and the T7 promoter needs at least 2 G for effective transcription.
(Note: If the specific target sequence already has 2 G, it is not necessary to add additional G, extra G will reduce the shear efficiency.)
- Specific sgRNA target sequence (20nt): the 5' terminal 20nt base sequence of target DNA sequence PAM (NGG).
- The 3' end of the primer: Scaffold template annealing sequence (14nt: GTTTTAGAGCTAGA).
Examples:
DNA template:
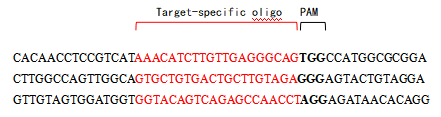
Forward primer (Target specific sgRNA oligo):

2.2 sgRNA synthesis
A. sgRNA template amplification
1) The reaction system is formulated according to the following system:
|
Components |
Volume(μL) |
Final concentration |
|
RNase free H2O |
Up to 25 |
- |
|
Scaffold Template |
1.25 |
- |
|
sgRNA Oligo(10 μM) |
1.25 |
0.5 μM |
|
2×Canace Enzyme Mix |
12.5 |
1× |
Note:
1. The Scaffold Template is pre-mixed with downstream primers.
- Use reagents and containers without RNase pollution.
2) PCR program:
|
Stage |
Temperature |
Time |
Cycles |
|
Denaturation |
98℃ |
10 s |
30 |
|
Annealing/Extension |
68℃ |
10 s |
|
|
Hold program |
4℃ |
∞ |
1 |
3) Agarose gel electrophoresis:
Using 2% agarose gel, 5 μL PCR reaction solution electrophoresis was used to detect the size and brightness of the bands, and 100bp DNA Ladder (Cat#10507ES) was used to calibrate the bands.
- In vitro transcription
- The reaction system is formulated according to the following system at room temperature::
|
Components |
Volume(μL) |
Final concentration |
|
RNase free H2O |
Up to 20 |
- |
|
10×sgRNA Reaction Buffer |
2 |
1× |
|
NTPs (25 mM each) |
8 |
10 mM each |
|
sgRNA tempate (from step A) |
5 |
- |
|
10×sgRNA Enzyme Mix |
2 |
1× |
Note:
1. Low temperature configuration may cause DNA template precipitation.
2. Use reagents and containers without RNase pollution.
2) Reaction program:
|
Temperature |
Time |
|
37℃ |
4 h |
|
4℃ |
∞ |
Note: The reaction is carried out in the PCR instrument, and the hot cover is opened to prevent the reaction liquid from evaporating for a long time.
3) DNA template digestion:
After the reaction was completed, 2μL DNaseⅠ (RNase free) was added to each tube and incubated at 37℃ for 15 min to remove the template DNA.
4) Pruduct purification:
Products can be purified using RNA Cleaner magnetic beads (Cat#12602ES), or phenol/chloroform purification method to remove proteins, free nucleotides, etc.
Notes
1. The reaction system must be strictly careful not to mix RNase.
2. RNase free experimental equipment (such as pipette and centrifuge tubes) are required.
3. Please wear the necessary PPE, such lab coat and gloves, to ensure your health and safety.
4. This product is for research use only.
Ver.EN20240710
Catalog No.:*
Name*
phone Number:*
Lot:*
Email*
Country:*
Company/Institute:*
Recommended products

