The classical reaction system is a one-time entry, and when the temperature rises from room temperature (25℃) to the maximum temperature (94±9 ℃), it may start a cyclic reaction that results in the synthesis of dimers. Under the influence of the Taq enzyme, the mismatch between the primer and the template as well as the two positions produced between the primers combine and extend numerous non-target sites. In subsequent PCR cycles, these non-specific products may continue to proliferate, leading to an accumulation of non-specific products while also depleting reaction system components and significantly reducing the particular products of the final PCR amplification. Therefore, to improve the specificity of PCR amplification and reduce the reaction's mismatch rate, the enzyme's activity can be artificially controlled. If the enzyme is not active before 72℃, it can avoid chain mismatch during the heating process (or configuring the reaction system), thereby ensuring amplification specificity. At present, the most commonly used method to improve the specificity of PCR is to use hot-start Taq DNA polymerase, that is, hot-start PCR.
1. Hot-start Taq DNA polymerase
2. Product features
3. Product performance
4. Ordering products
1. Hot-start Taq DNA polymerase
PCR is mainly composed of repeated thermal cycling of three steps high-temperature denaturation, low-temperature annealing, and suitable temperature extension. The template DNA is denatured into a single strand at high temperature, and the two primers anneal to a complementary sequence on the two template DNA strands under DNA polymerase and at a suitable temperature. Then, under the catalysis of DNA polymerase, four deoxynucleotide triphosphates (dNTPs) are used as substrates to extend the annealed primer, and then repeatedly, the DNA fragment located between the two known sequences exhibits geometric fold amplification.
Hot-Star PCR was developed to lessen or perhaps completely prevent these issues. Hot-start is a technique for improving the target amplification product in a PCR process while preventing non-target amplification. Primer dimer-induced mismatch or non-specific amplification can be significantly reduced by using DNA polymerase antibody inhibiting enzyme activity, which can render it inactive at low temperatures, denatured when heated to denaturing temperature, deblocking, and therefore releasing activity. The efficient detection of low-abundance genes is ensured by blocking antibodies, which prevent non-specific binding of DNA polymerase and template before predetermination and increase the efficiency of primers colliding with trace amounts of correct template annealing. These antibodies also significantly increase reaction sensitivity and amplification efficiency.
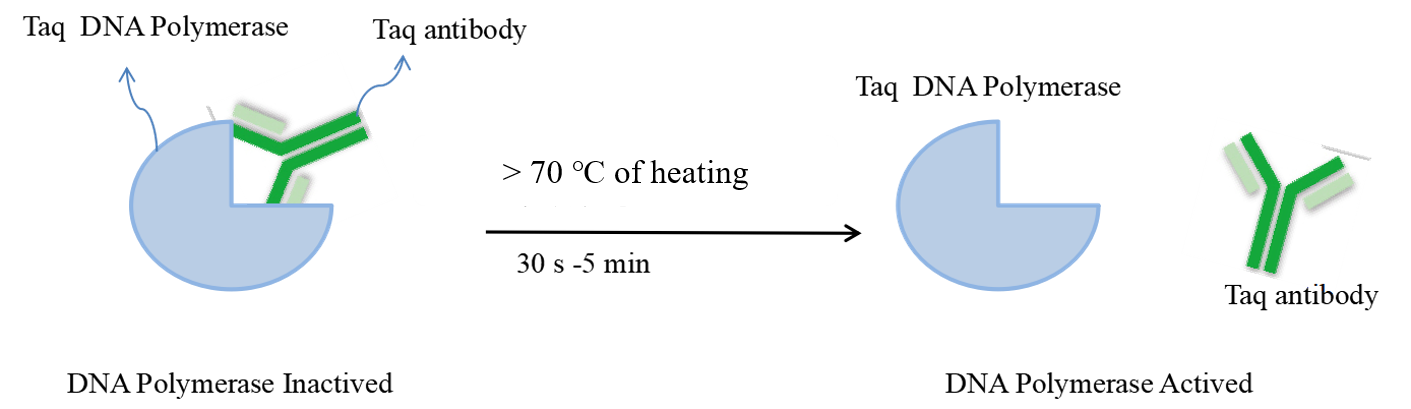
Figure 1:the principle of the hot-start enzyme
At present, the commonly used DNA polymerase hot-start modification methods on the market include chemical modification, ligand modification, and antibody modification. Different hot-start modification methods have certain differences in principle, and each has advantages and disadvantages.
1.1 Chemical modification method
The chemical modification of DNA polymerase generally uses small chemical molecules such as acid anhydrides and other organic substances to interact with the side chain amino acid (lysine) on the polymerase through chemical reactions. The enzyme loses its enzymatic activity when the temperature is low. When the temperature rises, the chemical bond between the acid anhydride and the amino group is broken, and the enzymatic activity is released, thereby achieving a hot start effect. Chemical modification can effectively block the activity of the enzyme. The operation is simple, but the release rate of the enzyme activity is slow. It depends on the temperature to activate the DNA polymerase and can effectively inhibit non-specific products. The reaction system can be configured at room temperature, and the PCR reaction buffer requirements are high.
1.2 Ligand method
Modification by ligand method mainly uses nucleic acid aptamers to block enzyme activity. As the name implies, nucleic acid aptamers generally refer to a class of single-stranded nucleic acid molecules with specific recognition functions, which can be either RNA or DNA, and are generally 25-60 nucleotides in length. As a recognition molecule of oligonucleotide sequence, it can target a wide range of molecules, from small molecules of inorganic metals to macromolecules in the biological field. These aptamers can form specific spatial conformations that are functionally similar in appearance to antibodies. It has high specificity and affinity for proteins or other small molecules. Nucleic acid aptamers bind to DNA polymerase through non-covalent bonds, thereby inhibiting the polymerization reaction of the polymerase at non-permissive temperatures. Generally, the screening of nucleic acid aptamers requires a short period, and the whole process can rely on automation, which is simple and fast.
1.3 Antibody method
Antibody-based hot-start enzymes generally consider the use of DNA polymerase antibodies to block the enzyme activity, making it inactive at low temperatures. When the antibody is denatured when heated to the denaturation temperature, the blocking is released to release the activity, which can greatly avoid non-specific amplification caused by mismatches or primer dimers. On the other hand, for low-abundance genes, blocking antibodies increase the efficiency of collision annealing between primers and a small amount of correct template while avoiding the non-specific binding of DNA polymerase and template before pre-denaturation. It ensures the effective detection of low-abundance genes and greatly improves the reaction sensitivity and amplification efficiency.
Table 1. Comparison of advantages and disadvantages of common modification methods for hot-start enzymes
|
Type of enzymatic blocking |
Chemical modification |
Ligand modification |
Antibody modification |
|
principle |
Small chemical molecules and DNA polymerase active center binding to block enzymatic activity |
Nucleic acid aptamer binds to polymerase through the non-covalent bond to block enzyme activity |
DNA polymerase antibody binds to polymerase to block enzyme activity |
|
Enzyme activity release conditions |
Temperature: greater than 95 ℃ Time: 10-15 min |
Temperature: greater than 60 ℃ Time: 1-5 min |
Temperature: more than 70 ℃ Time: 30 s-5 min |
|
advantage |
Hot-start modification allows more stringent conditions, maintains more stable enzymatic activity without any foreign DNA contamination, has strong specificity, and is cost-effective |
No activation step is required, high stability, reducing the possibility of sample degradation |
The antibody has no effect on the performance of the polymerase, the enzyme activation time is short, the enzyme activity is guaranteed, the affinity is strong, and the sensitivity is high |
|
shortcoming |
Longer enzyme activation time may affect enzyme activity, resulting in lower PCR product yields |
The nucleic acid ligand is a reversible inhibitor, not as stable as the chemical modification method, and the specificity is not strong enough |
Antibody modification is a kind of dynamic equilibrium, the production cost of antibody is high, and the optimization time of reagent ratio is long; the antibody of animal origin increases the risk of foreign genome contamination |
For the various hot-start methods described above, the chemical modifications cannot be synthesized in batches, and a long activation time is required before the polymerase can be released for activation. The effect of ligand-modified DNA polymerase is better at 20-25℃, and the polymerase activity is released at 40℃, and the specificity decreases. The antibody method modified hot-start enzyme has the best blocking effect, the release rate of enzyme activity is faster, and the PCR reaction time can be reduced. It is a hot-start enzyme method that is widely used in the IVD market at present.
2. Product features
Hot-start enzyme from Yeasen. is a mixture of Taq enzyme antibody (Cat. No. 31301ES) and Hieff™ D-Taq DNA Polymerase (Cat. No. 10709ES), the two of which have a high affinity. This product can be completely inactivated by heating it for 30 seconds at predenaturation temperature, which releases DNA polymerase activity. By using this hot-start Taq enzyme, it is possible to stop primer annealing-related amplification in its tracks. Both the 5 U/L and 50 U/L concentration items are dependably available from Yeasen.
👍1.Application in several circumstances: Using various Buffers, various IVD application scenarios are possible (such as genotyping, SNP typing, etc.)
👍2.Stable quality: Depend on a large-scale production system, a steady supply, and quick delivery for consistent quality.
3. Product performance
3.1 High sensitivity—— as low as 5 copies effectively detected(H3)
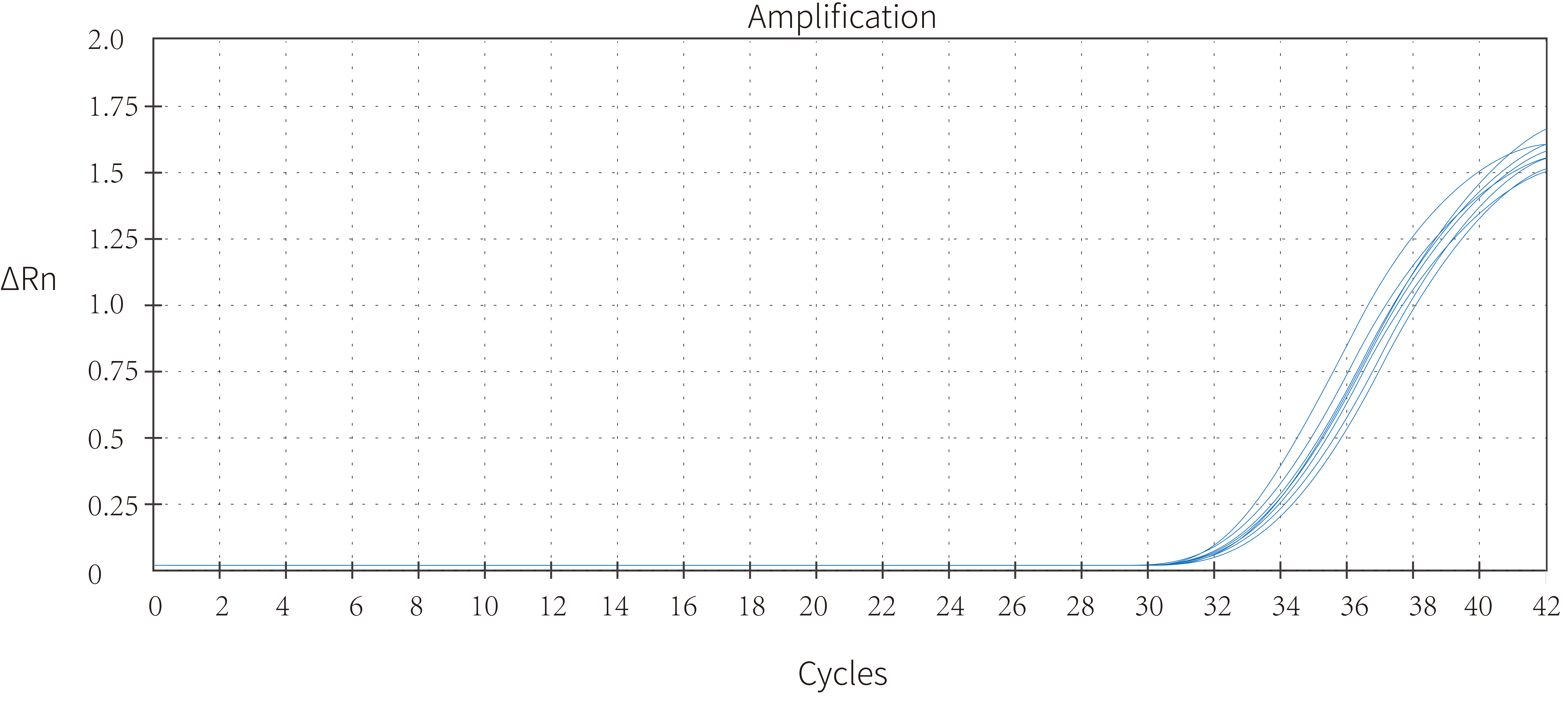
Figure 2. Five copies of the African swine fever plasmid were introduced using this product in the 25 μL reaction system, and eight rewells were detected with 100% accuracy. As a consequence, it was demonstrated that Hieff UNICON™ HotStart Direct Taq DNA Polymerase allowed for accurate detection of copies in the single-digit range.
3.2 Strong blood tolerance
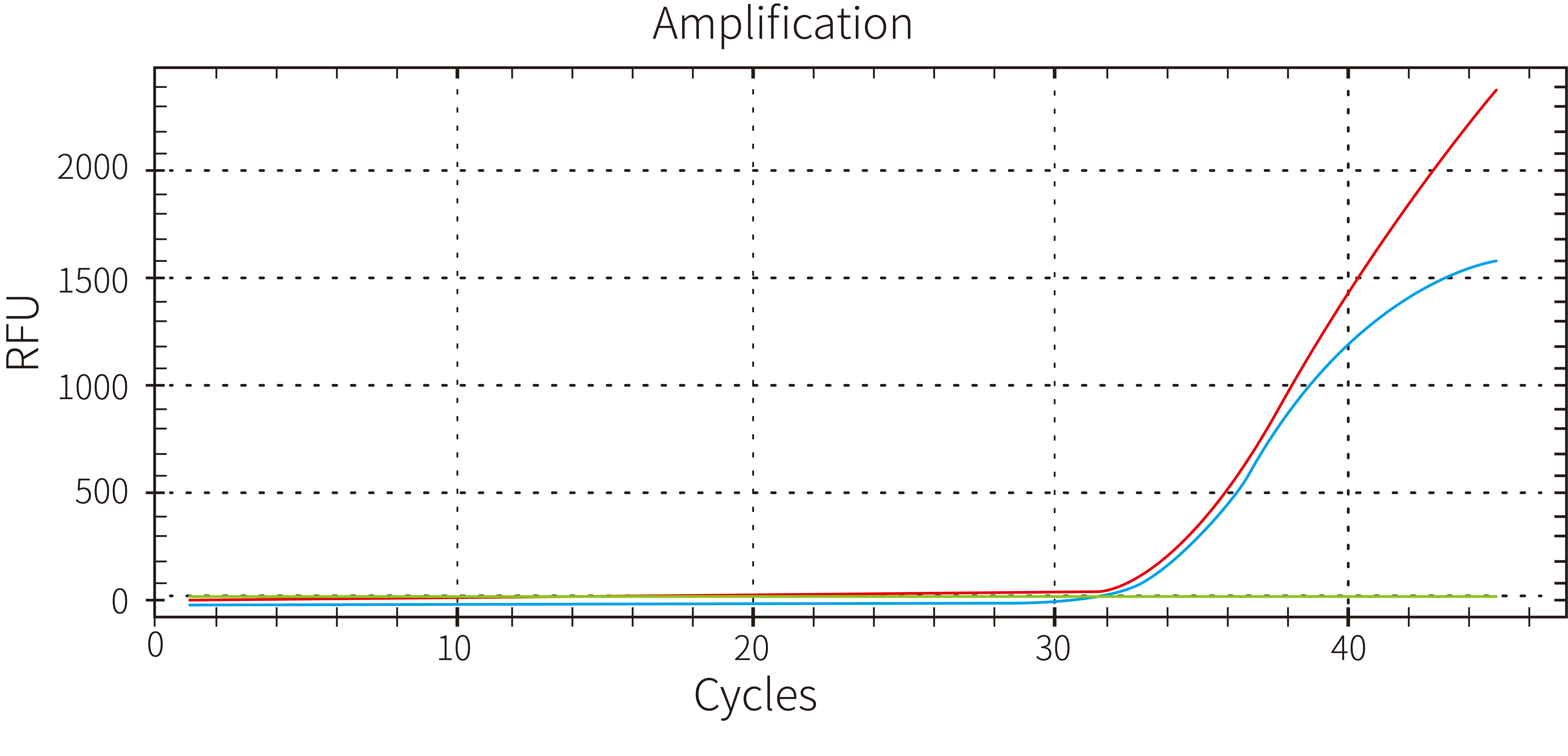
Figure 3. Use 5 μL of 40x diluted EDTA anticoagulant blood as an inhibitor additive in a 25 μL reaction system to identify 100 copies of the ASF virus. The outcomes demonstrated the blood tolerance of Hieff UNICON™ HotStart Direct Taq DNA Polymerase.
4. Ordering products
The ordering products provided by Yeasen are as follows:
Table 2. Ordering products
|
|
Product name |
Cat# |
|
Hot-start enzymes |
Hieff Union HotStart Direct Taq DNA Polymerase (5 U/μL) (Inquire) |
10717ES |
|
Hieff UNICON™ HotStart J-Taq DNA Polymerase (5 U/μL) (Inquire) |
10723ES |
|
|
10726ES |
||
|
Hieff UNICON Hotstart E-Taq DNA Polymerase (50 U/μL) (Inquire) |
10727ES |
References
[1] Michael R. Green, Joseph Sambrook. Hot Start Polymerase Chain Reaction (PCR)[J].Cold Spring Harbor Protocols, 2018, 2018(5).
[2] Kermekchiev, M. B . Cold-sensitive mutants of Taq DNA polymerase provide a hot start for PCR[J]. Nucleic Acids Research, 2003, 31(21):6139-6147. (IF19.16)
[3] Chou Q, Russell M, Birch D E, et al. Prevention of pre-PCR mis-priming and primer dimerization improves low-copy-number amplifications[J]. Nucleic Acids Research, 1992, 20(7):1717-1723. (IF19.16)