RNase R (20 U/μL)
Product description
Ribonuclease R (RNase R) from E. coli, is a magnesium-dependent 3′→5′ exoribonuclease that can digest all linear RNAs but does not digest lariat or circular RNA structures. Most cellular RNAs will be digested completely by RNase R, with the exception of tRNAs, 5S RNA and intron lariats. RNase R is used for enrichment of circRNAs in biological samples, identification of intronic lariat sequences, identification of exonic circRNAs, studying alternative splicing.
Features
Guaranteed efficacy and quality with ISO 13485 certification.
Available in bulk or tailored configurations.
Effectively breaks down linear RNA, while preserving lariat loops and circular RNAs.
Selective digestion of linear RNAs enhances the isolation of circular RNAs for protein production or intronic cDNA library construction.
Offering GMP-Grade RNAse R
Specificntions
| Source | A recombinant E. coli strain carrying the RNAse R gene from E. coli |
| Optimum Temperature | 37℃ |
| Storage Buffer | 50 mM Tris-HCl,100 mM NaCl, 0.1 mM EDTA, 0.1% Triton® X-100, 1 mM DTT, 50% glycerol,pH7.5 at 25℃ |
| Unit Definition | One unit converts 1 μg of poly-r(A) into acid-soluble nucleotides in 10 minutes at 37°C in 20 mM Tris-HCl (pH 8.0), 100 mM KCl and 0.1 mM MgCl2. |
Application
RNase R serves as a fundamental component in circular RNA synthesis (circRNA). CircRNA holds promise for enhancing pharmaceutical stability and biostability. With their covalently closed structures, circRNAs withstand RNA exonucleases. [1,2] RNase R finds widespread application in various studies, including alternative splicing, intron cDNA production, and the isolation of splicing intermediates and lariats.
Components
|
Components No. |
Name |
14606ES72 |
14606ES86 |
14606ES92 |
|
14606-A |
RNase R (20 U/μL) |
12.5 μL |
125 μL |
500 μL |
|
14606-B |
10×RNase R Reaction Buffer |
250 μL |
3 × 1 mL |
10 mL |
Storage
This product should be stored at -25~-15℃ for 2 years.
Instructions
Prepare the following reaction system in a sterile microcentrifuge tube
|
component |
Volume |
|
10×RNase R Reaction Buffer |
2 μL |
|
RNA sample |
1 μg |
|
RNase R (20 U/μL) |
2-4 U |
|
RNase-free ddH2O |
Up to 20 μL |
Incubate at 37 ℃ for 10 - 30 minutes.
Figures
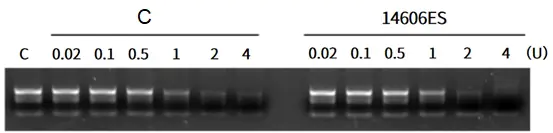
Figure 1: Linear RNA molecules can be digested by Yeasen RNAse R.
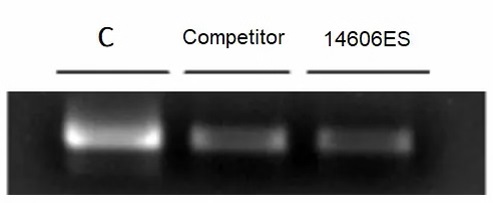
Figure 2: CircRNA can not be digested by Yeasen RNAse R.
Notes
1. The activity of RNase R requires 0.1-1.0 mM Mg2+;
2. With the increase of RNA, the reaction time can be extended and the enzyme amount can be increased appropriately;
3. The EDTA content in RNA samples may affect the activity of RNase R.
4. Incubate at 70 ℃ for 10 minutes can inactivate the enzyme.
Reference
1. Qu L, Yi Z, Shen Y, et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell. 2022;185(10):1728-1744.e16. doi:10.1016/j.cell.2022.03.044
2. Chen L, Wang C, Sun H, et al. The bioinformatics toolbox for circRNA discovery and analysis. Brief Bioinform. 2021;22(2):1706-1728. doi:10.1093/bib/bbaa001
Catalog No.:*
Name*
phone Number:*
Lot:*
Email*
Country:*
Company/Institute:*

